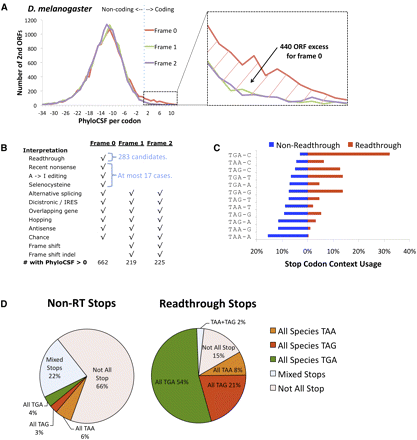
Evidence of readthrough mechanism. (A,B) Excess of high-scoring regions in-frame (frame 0) compared to out-of-frame (frame 1, frame 2) suggests readthrough as the likely mechanism and provides an estimate of readthrough count. (A) PhyloCSF score per codon (x-axis) of the regions starting 0, 1, or 2 bases after all D. melanogaster annotated stop codons (red, green, purple, respectively) and continuing until the next stop codon in that frame, excluding regions that overlap another annotated transcript. Frame 0 shows an excess of more than 400 predicted protein-coding regions compared with the other reading frames, suggesting abundant readthrough. In contrast, a similar plot for Caenorhabditis elegans shows no significant excess in frame 0 (Supplemental Fig. S11), suggesting that the abundance of readthrough in Drosophila is not universal. (B) Possible mechanisms associated with protein-coding function downstream from D. melanogaster stop codons (rows) and associated reading frame offsets where corresponding protein-coding function is expected (columns). Random fluctuations would lead to an even distribution among the three frames, as would unannotated alternative splice variants and unannotated IRESs (note that annotated splice variants and IRESs have already been excluded), while frameshift events and recent frameshifting indels would bias away from frame 0. A bias for in-frame protein-coding selection is expected only for stop codon readthrough, recent nonsense mutations, A-to-I editing, and selenocysteine, the latter three together accounting for at most 17 cases. This leaves readthrough as the only plausible explanation for an excess of ∼420 frame 0 regions with positive PhyloCSF scores. (C) Usage of stop codon context (stop codon and subsequent base) provides additional evidence of a readthrough mechanism. The 4-base contexts are sorted in order of decreasing frequency among the 14,928 non-readthrough stop codons (blue), with less frequent stop codons (top, e.g., TGA-C) experimentally associated with translational leakage in other species and most frequently associated with efficient termination (bottom, e.g., TAA-A). Context frequencies for readthrough candidates (red) are opposite of non-readthrough transcripts, suggesting a preference for leaky context, with one-third using TGA-C and almost none using TAA-A. (D) Increased stop codon conservation in readthrough candidates. Only ∼1/3 of D. melanogaster non-readthrough stop codons have aligned stops in all 12 species, and only ∼1/3 of those are perfectly conserved (i.e., have the same stop codon in all 12 species). In contrast, 83% of candidate readthrough stop codons have an aligned stop in all 12 species, and 97% of those are perfectly conserved. While all three stop codons are involved in readthrough of different genes, individual readthrough genes rarely show substitutions between different stop codons, suggesting that the three stop codons are not functionally equivalent. Moreover, the only eight substitutions observed are between TAA and TAG, with no substitutions involving TGA, even though it is the most frequent readthrough stop codon, suggesting that TAA and TAG are functionally similar.











