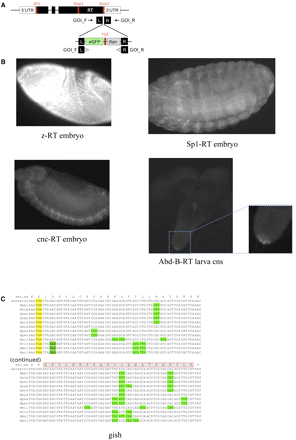
Experimental validation of readthrough. (A) GFP insert construct replacing the second stop codon so that GFP is only observed after translation of the 3′ end of the second ORF and subsequent eGFP gene. GOI_F and GOI_R are 50-bp homology arms on the forward and reverse strands specific to each gene of interest (GOI). (B) Expression of GFP in transgenic constructs showing that translation continues through to the second stop codon for four of the readthrough (RT) candidates. Strains shown are z-RT, Sp1-RT, and cnc-RT in embryos, and Abd-B-RT in the central nervous system of a larva. No GFP expression was found in a wild-type strain used as a control (Supplemental Fig. S12). (C) Mass spectrometry evidence of readthrough. Example of readthrough region (gish) supported by a 22-amino-acid peptide match (red rectangle) to mass spectrometry Drosophila PeptideAtlas (one of nine cases). With no ATG codon between the stop codon and the peptide, and no observed alternative splicing events across thousands of RNA-seq reads overlapping this region, readthrough seems the only plausible explanation for translation of this peptide.











