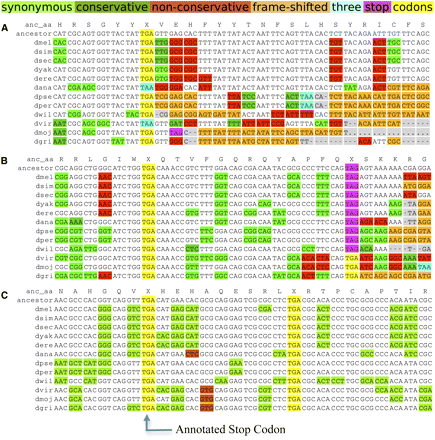
Protein-coding evolutionary signatures for typical, readthrough, and double-readthrough stop codons. Alignments surrounding the annotated stop codons of three genes for 12 Drosophila species and their inferred maximum-parsimony common ancestor. The color coding of substitutions and insertions/deletions (indels) relative to the common ancestor is a simplification for visualization purposes, as the actual PhyloCSF score sums over all possible ancestral sequences and weighs every codon substitution by its probability. Insertions in other species relative to D. melanogaster are not shown. (A) Alignment of a typical gene (bw), shows abundant synonymous and conservative substitutions (green) upstream of the stop codon, and many non-conservative substitutions (red), frameshifting indels (orange), and in-frame stop codons downstream from the stop codon. The stop codon locus shows several substitutions between different stop codons. (B) Alignment of CG17319, one of 283 readthrough candidates. The region between the annotated stop codon and the next in-frame stop codon shows mostly synonymous substitutions and lacks frameshifting indels, while the region downstream from the second stop shows non-conservative substitutions and indels typical of non-coding regions, providing evidence of continued protein-coding selection in the region between the two stop codons, and suggesting likely translational readthrough of the first stop codon. As is typical for readthrough candidates, the first stop codon is perfectly conserved, while the second stop codon shows substitutions between different stop codons. (C) Alignment of a double-readthrough candidate, Glu-RIB (one of 16 cases). Both the second ORF and the third ORF show protein-coding signatures, indicating that both stop codons are likely readthrough. Both readthrough stop codon positions show no substitutions.











