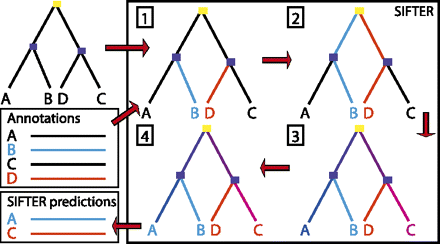
Overview of SIFTER method. (1) A reconciled protein phylogeny and a file containing some of the family's proteins with annotations (blue and red functions) are input into SIFTER. SIFTER incorporates the observations from the annotation file into the phylogeny. (2) Message passing propagates the observations from the leaf nodes to the root of the tree. (3) Message passing then propagates information from the complete set of observations back down to the leaves of the tree, enabling extraction of posterior probabilities at every node in the tree. Intuitively, posterior probabilities at a node take into account every observation scaled by the tree distance. (4) Predictions can be extracted from the tree using, in our case, the function with the maximum posterior probability for each protein.











