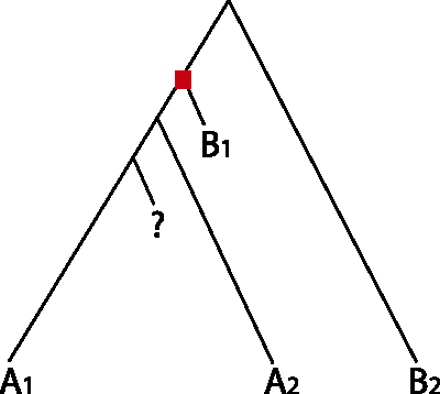
Sequence similarity does not directly reflect phylogeny. The proteins in this tree have either molecular function A or B. There is a duplication event indicated by a red square and the query protein is denoted by “?”. The most significant BLAST hit for the query protein will be B1 because the path length in the phylogeny from the query protein to B1 is the shortest. Thus, BLAST-based prediction methods will transfer B to the query protein. However, it is more likely that the tree has only one functional mutation, in which ancestral function B mutated to function A on the left-hand side of the bifurcation after the duplication event. So A is a better annotation for the query protein. A phylogenetics-based approach reaches this conclusion naturally. Example adapted from Eisen (1998).











