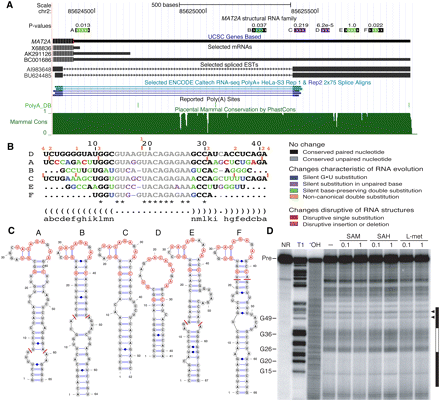
Family of hairpins in 3′-UTR of MAT2A. (A) Location of the six hairpins (named A–F) of the MAT2A 3′-UTR family. The initially predicted UTRP family consists of C, D (EvoFold predictions, purple), and B (paralog search hit, dark green). Hairpins A, E, and F were found by a dedicated, more lenient search for paralogs (light green). The well-conserved core part of the hairpins can be extended in some cases (black flanks). A putative 3′-UTR alternative intron is indicated by spliced EST and RNA-seq evidence. (B) Color-coded alignment of the human sequence from all family members. The alignment is referenced by D. Location and length of relative insertions in the other sequences are indicated (orange bars and numbers). The loop region reveals a motif of bases that are completely conserved among all six members (*). (C) Structures of all six extended hairpins showing the boundary of single sequence predictions (red bars) as well as the fully conserved motif (red nucleotides). Note that hairpin D can also form the two base pairs of the loop regions seen for the other hairpins, although not predicted by EvoFold. (D) In-line probing analysis of the 186-long MAT2A construct (including hairpin A). RNA cleavage products resulting from spontaneous transesterification during incubations in the absence (−) of any candidate ligand or in the presence of SAM, S-adenosylhomocysteine (SAH), and L-methionine (L-met), each tested at concentrations of 0.1 mM and 1 mM, were resolved by denaturing 10% PAGE. (NR) No reaction; (T1) partial digest with RNase T1; (−OH) partial alkaline digest; (Pre) precursor RNA. Selected bands in the T1 lane are labeled with the positions of the respective 3′-terminal guanosyl residues, according to the numbering used for hairpin A in panel C. Filled bars correspond to positions within hairpin A that are predicted to be largely base-paired, while the open bar corresponds to positions within the putative loop sequence. (Arrowheads) Putative bulged nucleotides C50 and A55.











