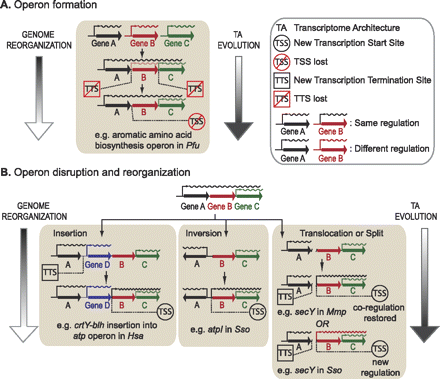
The role of parallel evolution of TA in buffering intermediate genome organization states during formation, disruption, and reorganization of operons. (A) Operon formation. The assembly of functionally linked genes occurs through genome organizational events that bring them into close proximity. Parallel evolution of the TA via spontaneous mutations modifies transcriptional signals in the form of generating new transcriptional elements (TSSs or TTSs) or killing existing elements to generate polycistronic transcripts. Some internal transcriptional elements are retained and used in a conditional manner (i.e., triggered or silenced as a function of environmental context) to accommodate alternate regulatory schemes in certain environments. Needless to say, all of these events are driven by natural selection as genome streamlining [assembly of genes into operons to increase coding density and deletion of unnecessary genomic elements (coding and noncoding)] is associated with gain of fitness over competition. (B) Operon disruption and reorganization. While the random nature of genome shuffling allows organisms to continually explore novel fitness landscapes in changing environments, it can also be detrimental as it results in disruption of existing operon structures more often than yielding new beneficial arrangements. The parallel evolution of TA continues to buffer the intermediate genome states by restoring coordinate transcriptional control of functionally linked genes (indicated in the examples illustrating insertion and inversion events). However, occasionally this dynamic process allows for the evolution of new regulatory schema as illustrated in the second example of a translocation or split event. We have provided specific examples as evidence to support the proposed model.











