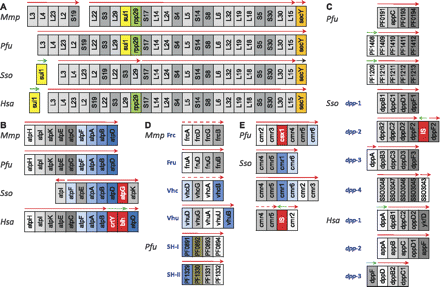
Reorganization events within highly conserved operons. Operons for ribosomal proteins (A), ATP synthase (B), oligo/dipeptide transporter (C), hydrogenase (D), and RAMP module Cas proteins (E). Homologous genes are shown in the same color; red shaded genes do not have orthologs in the other three archaea. Arrows above the operons indicate direction and span of TUs determined in this study. The different arrow colors represent different expression patterns. For instance, secY in Mmp is transcribed as a monocistronic TU that is co-expressed with the large ribosomal operon. In contrast, secY in Sso is also transcribed as a separate TU, but its expression pattern is different from that of the large ribosomal operon. Dotted arrows indicate unexpressed ORF(s).











