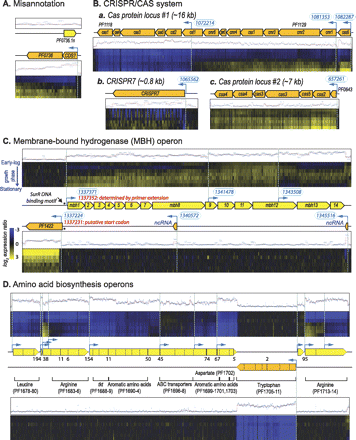
Examples of dynamic changes in transcriptome architecture of P. furiosus DSM 3638. (A) Identification of misannotation. While there was no transcription of the hypothetical protein-coding gene PF0736.1n (P_expressed = 0.16), a transcript was detected from the opposite strand and assigned to a new putative gene encoding a 74-amino acid (aa)-long protein. It should be noted that PF0736.1n was previously believed to be a bona fide gene based on RNA hybridization to a PCR-based double-stranded microarray (Poole et al. 2005). This example showcases the value of strand-specific analysis. (B) CRISPR/CAS system. In the Pyrococcus CRISPR-Cas system, the guide crRNAs were suggested to be processed and translocated to the Cmr complex by the ribonuclease Cas6. (a) We detected separate TUs for each cas6 and cmr gene cluster. (b) The relative change in these transcripts was highly correlated with changes in all seven CRISPR elements and a large number of computationally predicted small nucleolar RNAs (snoRNAs) (r > 0.9). Unexpectedly, the adjacent core cas genes (cas1, cas4, cas5t, cas6) and other cas genes (cst1 and cst2) had different transcriptional profiles, suggesting conditionally activated transcriptional elements within this operon. (c) While the core cas gene cluster at locus #1 was down-regulated throughout growth in batch culture, the cas genes at locus #2 were up-regulated (r ∼ 0.4). (C) Conditional regulation of MBH operon. Segmentation analysis identified three alternative TSSs: The first was located upstream of mbh1-9, an operon that encodes subunits of a putative Na+/H+ antiporter; the second was located upstream of mbh10-12 (hydrogenase 3 complex subunits [hycG and hycE]), and the third was located upstream of mbh13-14 (hycD and hycF). Interestingly, these TUs separate the transcription of membrane-associated proteins encoded by Mbh1-9 from the cytoplasmic proteins encoded by Mbh10-12 (Holden et al. 2001). We detected at least two ncRNAs that were antisense to the MBH operon; the ncRNA located at the 3′ end of the MBH operon was correlated with surR (r = 0.95). The location of the TSS for mbh1 detected by the segmentation analysis mapped to within 19 nt of the TSS that was previously determined by primer extension (Lipscomb et al. 2009). We also observed that the TSS for PF1422 was located 40 nt inside the coding sequence; notably, there is a start codon immediately internal to this TSS, suggesting that the originally assigned start codon for this gene is incorrect. (D) Amino acid biosynthesis operons. A distinguishing feature of Pfu is the clustering of amino acid biosynthesis operons (leucine, arginine, aromatic amino acids, tryptophan) in a contiguous stretch (34 kb, 1560860–1594957). Genes in the forward and reverse strands are shown in yellow and orange, respectively. Gene boundary is indicated with a dotted line when adjacent genes are overlapping and with a solid line if there is space between genes. Numbers below or above the arrows denote the positive intergenic distances. See Figure 2 for additional keys to interpreting notations in this figure. Examples of dynamic changes in TA of Sso can be found in Supplemental Material, and TA of Hsa has been reported in our previous publication (Koide et al. 2009).











