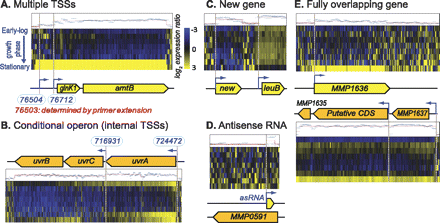
Examples of dynamic changes in transcriptome architecture of M. maripaludis S2. The tiling array data were plotted against coordinates on the genome, and transcriptional units discovered by the automated segmentation approach were manually inspected and curated through interactive exploration in the Gaggle Genome Browser (Bare et al. 2010). Genes in the forward and reverse strands are shown in yellow and orange, respectively. Corresponding transcriptome architecture (TA) data are aligned above forward strand genes and below reverse strand genes. The blue horizontal bars represent probe intensity (log2 scale) at the corresponding genomic location for reference RNA, which was prepared from a mid-log phase culture. The overlaid red line is a model fit by a segmentation algorithm that was applied to determine breaks in transcript signals (i.e., TSSs and TTSs). The heat map indicates transcript level changes at eight time points over various phases of growth in batch culture ratios (log2 scale) relative to reference RNA (blue is down-regulated; yellow up-regulated). (A) Multiple TSSs. Transcription is initiated at two sites (blue bent arrows) upstream of the glnK1-amtB operon, which encodes nitrogen regulatory protein P-II and an ammonium transporter, respectively. Interestingly, one of these TSSs (76504) was discovered using primer extension, and the TSSs mapped by the two independent methodologies mapped within one nucleotide of each other. This example illustrates the power of global analysis in comprehensive analysis of TA. (B) Conditional operon. Analysis of predicted operon structures identifies unexpected conditional breaks in the organization of the operon during cellular responses in differing environments. The mechanisms for a broken operon could include conditional activation of internal promoters or terminators, or conditional cleavage and processing. We show one example of a conditional operon for three DNA repair genes uvrABC. (C) Discovery of a new gene. We have discovered at least 63 transcripts in genomic locations that were not assigned to any annotated features. Here, we show an example of a newly discovered transcript that encodes a protein homologous to a hypothetical protein from Methanococcus maripaludis C6 (E-value = 2 × 10−13). (D) Discovery of an antisense ncRNA. At least 28 antisense ncRNAs were discovered. The example shown is for an ncRNA that is antisense to the 5′ end of MMP0591. (E) Discovery of fully overlapping genes. We have identified transcription of the antisense strand of MMP1636 encoding a major facilitator transporter. This newly discovered gene is interspersed between and cotranscribed with MMP1635, a redox-active disulfide protein, and MMP1637, a hypothetical protein.











