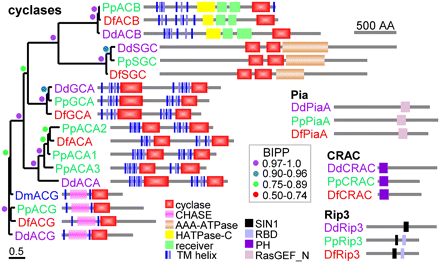
Architectural conservation of adenylate and guanylate cyclases. Proteins were identified by TBLASTN search with DD cyclase sequences and query of SACGB with the IPR001054 A/G_cyclase domain. Protein phylogenies of nucleotidyl cyclases were constructed using Bayesian inference (Ronquist and Huelsenbeck 2003) from alignments of the shared functional domains as described in the legend to Supplemental Figure S5.6. For proteins with two cyclase domains, a tandem alignment of both domains was used, with the single domains of ACB and ACG used twice. The tree is presented unrooted and decorated with the domain architectures of the proteins as analyzed by SMART (Schultz et al. 1998). The posterior probabilities (BIPP) of nodes are indicated by colored circles. Locus and gene ids: (ACB) DD: DDB_G0267376, PP: PPL_10642, DF: DFA_10857; (SGC) DD: DDB_G0276269, PP: PPL_05930, DF: DFA_12195; (GCA) DD: DDB_G0275009, PP: PPL_11010, DF: DFA_10832; (ACA1) DD: DDB_G0281545, PP: PPL_01657, DF: DFA_04345; (ACA2) PP: PPL_12370; (ACA3) PP: PPL_10658; (ACG) DD: DDB_G0274569, PP: PPL_06767, DF: DFA_08118; (Pia) DD:DDB_G0277399, PP: PPL_02311, DF: DFA_00057; (CRAC) DD: DDB_G0285161, PP: PPL_07100; (Rip3) DD: DDB_G0284611, PP: PPL_11127, DF: DFA_06425.











