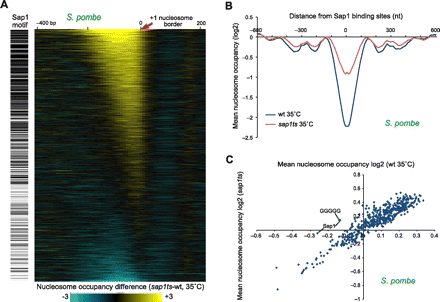
Sap1 acts as a GRF in S. pombe. (A) Sap1 deletion in S. pombe results in increased nucleosome occupancy at Sap1 motifs. (Left) Genes with significant matches to the Sap1 binding half-site (black). (Right) Difference in nucleosome abundance at each gene in S. pombe (rows) between sap1ts and wild-type strains (both at restrictive temperatures). Genes are aligned by the +1 nucleosome/NFR boundary (0, red arrow) and ranked from gain (top, yellow) to loss (bottom, blue) in nucleosome occupancy over their NFR. (B) Increased nucleosome occupancy at Sap1 half-sites in a sap1ts strain in S. pombe. Shown are log2 nucleosome occupancy averages at all genes with a significant Sap1 motif match in their upstream promoter for a sap1ts strain (red) and a wild-type strain (blue) grown in restrictive temperatures (35°C). Genes are aligned by the location of the Sap1 motif. Nucleosome occupancy increases over Sap1 sites is characteristic of GRF activity. (C) Increased nucleosome occupancy in 5-mers, reflecting the Sap1 half-sites in a sap1ts strain compared with wild-type (both at restrictive temperature, 35°C). Shown is the mean nucleosome occupancy (log2) for each 5-mer in the wild-type (x-axis) and the sap1ts strain (y-axis). Sap1 half-sites are labeled. The only additional site with increased occupancy is the intrinsic sequence GGGGG.











