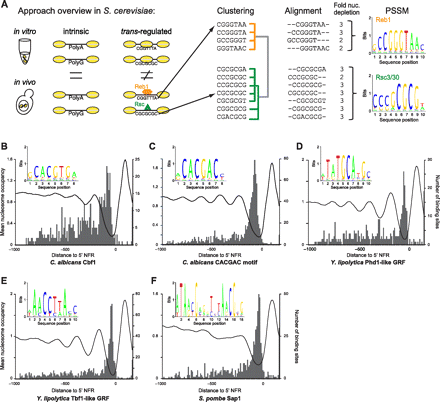
Identification of novel general regulatory factor (GRF) motifs. (A) Overview of GRF motif discovery approach. Nucleosome-depleted sequences are first classified as intrinsic (left) if they evict nucleosomes in vitro and in vivo, or as trans-regulated (right) if they evict nucleosomes more strongly in vivo. Trans-regulated nucleosome positioning sequences in each genome are then clustered based on similarity, aligned, and combined into a position specific scoring matrix (PSSM). Shown are the results for S. cerevisiae, where the algorithm outputs the known PSSMs of chromatin regulators Reb1 and Rsc3/30. (B–E) Predicted binding sites for GRFs in C. albicans (B,C), and Y. lipolytica (D,E). Shown are the sequence logos of the PSSMs (insets) for the sites learned by our approach from 7-mers depleted in each species. For Y. lipolytica, the names of S. cerevisiae proteins with similar sequence specificity are displayed. Each graph shows the average normalized nucleosome occupancy (left y-axis) in promoters with significant matches to the PSSM (black curve, aligned by a gene's +1 nucleosome), as well as the location (gray bars) and number (right y-axis) of binding-site locations. As observed with GRFs in other species (Tsankov et al. 2010), these binding sites are almost entirely (>89%) NFR-localized. (F) Known Sap1 motif (Ghazvini et al. 1995) is identified as a GRF site in S. pombe. As in B–E but for S. pombe data.











