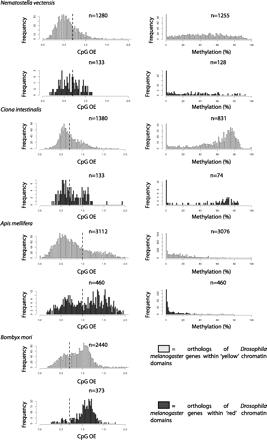
Gene-body CpG content and methylation profiles of YELLOW and RED gene orthologs. Plotted are frequency distributions of gene-body CpG density (left panels) and DNA methylation (right panels). The CpG density cut-off between low-CpG and high-CpG genes in the four organisms (0.7 for N. vectensis, C. intestinalis, and B. mori; 1.0 for A. mellifera) is indicated by a vertical dashed line. YELLOW gene orthologs (light gray) display significantly lower gene-body CpG density and higher gene-body CpG methylation compared with RED gene orthologs (dark gray) in all cases (P < 0.005 for all except C. intestinalis gene-body CpG density, P = 0.034, Wilcoxon rank sum tests with continuity correction). The DNA methylation analysis could not be performed for B. mori due to the very small number of methylated genes.











