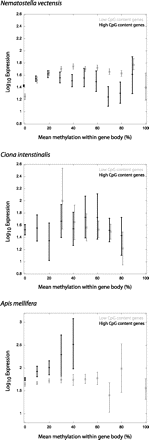
Expression levels of low- and high-CpG genes. Plotted are log10 RPKM values obtained from Zemach et al. (2010). The x-axis represents the average expression values within 10% methylation bins spanning 0%–100%. Error bars, SD. For A. mellifera, there were very few data points available for high-CpG genes with >40% gene-body methylation. Plotted are 9442 low-CpG and 12,116 high-CpG genes (N. vectensis), 448 low-CpG and 561 high-CpG genes (C. intestinalis), and 3077 low-CpG and 2838 high-CpG genes (A. mellifera). (Light gray) Data for low-CpG genes; (dark gray) high-CpG genes.











