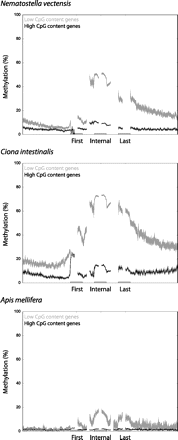
Exonic methylation peaks are prominent in low-CpG genes. DNA methylation values were calculated for 2-kb windows centered on the transcriptional start site, “internal” exons (i.e., not first or last exons), and last exons. Trend lines represent the median methylation values for all genes within that category for which data were available, and vertical, faint gray lines show the standard deviations. (Light gray) Low-CpG genes; (dark gray) high-CpG genes. The trend lines are interrupted because exon sizes vary within any genome, and it is not straightforward to generate a composite plot that accurately represents methylation profiles over the entire exon. Our plots show methylation levels 100 bp upstream of and downstream from exon–intron junctions. Raw BS-seq data are from Zemach et al. (2010). Number of genes plotted: 10,827 low-CpG and 14,566 high-CpG genes (N. vectensis), 3821 low-CpG and 4336 high-CpG genes (C. intestinalis), and 4064 low-CpG and 4446 high-CpG genes (A. mellifera). High-CpG genes in C. intestinalis seem to be associated with a small peak of methylation at the transcriptional start site (TSS). However, this is simply due to the cut-off between low and high-CpG genes not being perfect; that is, the high CpG gene category is likely to contain some low CpG genes. Also, incorrect genomic annotation (i.e., incorrectly marked TSSs) is also likely to have an influence. None of this appreciably affects our overall conclusions since the vast majority of the “signal” is from the appropriate gene category.











