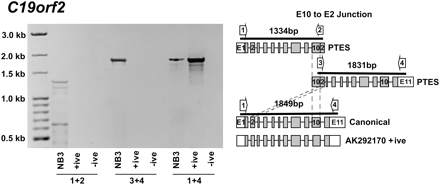
Identification of extended PTES products using junction-specific primers. Amplicons generated using primers specific for C19orf2 PTES E10-E2 splice junction are shown. Primers 1 and 2 amplify from the 5′ UTR to the E10-E2 breakpoint. Primers 3 and 4 amplify from the E10-E2 breakpoint to the 3′ UTR. Templates are as follows: (NB3) cDNA where the E10-E2 PTES product was originally identified; (+ive) unrearranged (canonical) C19orf2 cDNA clone (AK292170); and (-ive) no template. The exon organization of the inferred E10-E2 spliced PTES RNAs, the full-length C19orf2 gene from RefSeq (canonical), and the AK292170 +ive control is also shown, together with the position of primers used and the expected amplicon sizes. Individual exons are shown as boxes, with coding regions shown in gray and UTRs in white. The sizes of the PTES amplicons are given relative to the E10-E2 junction. Additional products of ∼0.4–0.65 kb and ∼1.2 kb are seen when the NB3 template is amplified using primers 1 and 2, suggesting that shorter C19orf2 PTES isoforms also exist. For all primers, see Supplemental Table S6.











