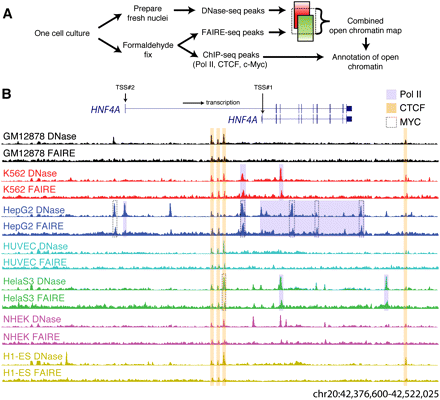
Identification of open chromatin in seven human cell lines. (A) A schematic representation of the experiment and analysis design. (B) DNaseI (y-axis fixed at Parzen signal value 0.15) and FAIRE (y-axis fixed at 0.04) data from seven cell lines surrounding the HNF4A locus (145 kb; UCSC Genome Browser) shows both ubiquitous and cell-type selective open sites that are especially prevalent in HepG2 cells. Pol II, CTCF, and MYC ChIP-seq peaks that overlap open chromatin are highlighted.











