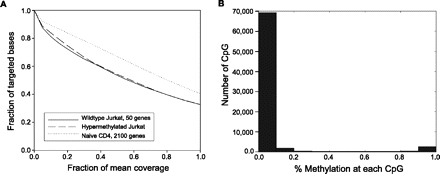
(A) Coverage summary for targeted BiS-seq. The fraction of bases with coverage greater than the specified fraction of mean read depth is plotted for 50-promoter assay in Jurkat wild-type (solid line; mean read depth = 17,011), Jurkat hypermethylated (dashed line; mean = 17,274), and 2,000-promoter assay in primary naive CD4 T cells (dotted line; mean = 1,320). The CD4 assay included amplicon concatenation before shearing, which increased coverage at 50% of mean read depth to 67% of the targeted amplification region. (B) Bimodal distribution of methylation across all assayed CpG.











