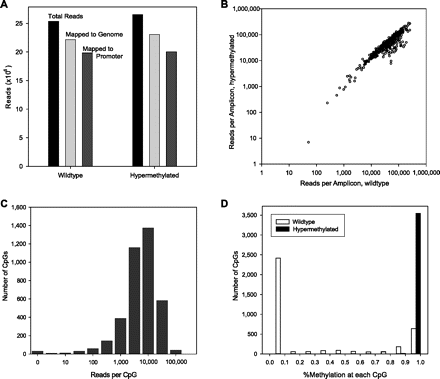
Effectiveness of custom primer design and highly multiplexed PCR. (A) Targeted loci in wild-type and methyltransferase-treated (hypermethylated) Jurkat DNA were amplified, sequenced, and mapped with Novoalign. Over 85% of reads mapped to the genome, and a similar fraction of mapped reads were uniquely mapped to amplicon regions. (B) Custom primers yielded generally high read counts across 393 amplicons and minimal methylation-induced bias. (C) High read depth at each CpG allows accurate measure of methylation state and suggests that greater multiplexing can be applied. (D) Bimodal (wild-type) or unimodal (hypermethylated) distribution of methylation across all assayed CpG.











