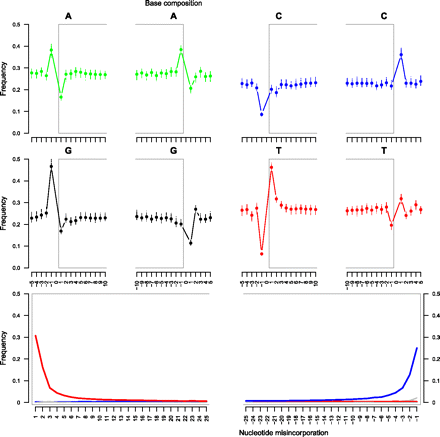
Illumina sequencing: DNA fragmentation and nucleotide misincorporation patterns on ancient horse reads. (Top, middle) The base composition of the reads is reported for the first 10 nucleotides sequenced (left: 1–10) as well as for the five nucleotides located upstream of the genomic region aligned to the reads (left: −5 to −1). In addition, the base composition of the last 10 nucleotides sequenced (right: −10 to −1) and of the five nucleotides located downstream from the reads (right: 1–5) in the genome equCab2 is provided. Nucleotide positions located within reads are reported with a gray frame. Each dot reports the average base composition per position as estimated from reads mapping against chromosomes 1–31 and X. The range of the base composition per individual chromosome is also reported. (Bottom) The frequencies of all possible mismatches and indels observed between the horse genome and the reads are reported in gray as a function of distance for 5′- to 3′-ends (first 25 nucleotides sequenced) and 3′- to 5′- (last 25 nucleotides), except for C→T and G→A, which are reported in red and blue, respectively. The latter variations range from 0.6% to 30.7% per site (5′- to 3′- end) or 0.7%–25.1% per site (3′- to 5′- end) and exceed the variations observed for other misincorporation types that are consequently mostly hidden in the figures (<0.1%–0.9% per site). The misincorporation frequencies are calculated by dividing the total number of occurrences of the modified base at a given position in a read by the total number of the unmodified base at the same position in the horse genome.











