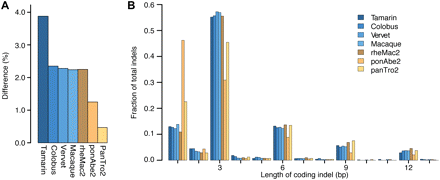
Sequence differences and indel lengths in protein-coding regions. (A) Coding sequence differences relative to the human reference genome for each assembled exome and non-human primate reference genome, calculated from the 9,106,235 sites that are high-quality in all species. (B) Distribution of coding indel lengths from the 4637 gene alignments where at least 75% of sites have high-quality sequence in all species. All indels are relative to the human reference genome. Low-quality indels are not included unless their read depth is ≥4 or they are confirmed by a high-quality indel in another species. Lengths from indels <15 bp apart are combined to account for uncertainty in the alignments. Indel lengths from the exome assemblies of macaque, vervet, colobus, and tamarin (blue); indel lengths from the reference genomes of chimpanzee (panTro2), orangutan (ponAbe2), and macaque (rheMac2) (yellow).











