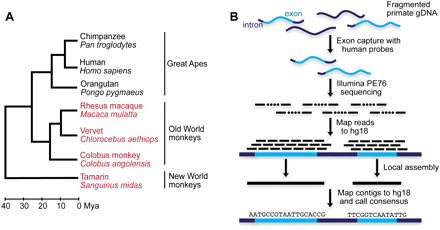
Sequence capture and assembly of non-human primates. (A) Phylogeny of primate species used in all analyses. Sequences from the species in red and black are from our exome assemblies and the publicly available primate reference genomes, respectively. For rhesus macaque, we generated an exome assembly and compared it to the macaque reference genome to assess the accuracy of our capture and assembly method. The phylogenetic relationship of the species and estimates of their divergence dates are from Goodman (1999). (B) Overview of sequencing, mapping, and assembly pipeline. Primate genomic DNA is fragmented, and protein-coding regions are captured using a solution-based hybridization method and sequenced as 76-bp paired-end reads. Reads are mapped to the repeat masked human reference genome using cross_match (v1.090518, http://www.phrap.org). Reads with overlapping mapped chromosomal coordinates are partitioned into groups and assembled independently using phrap (v1.090518, http://www.phrap.org). The resulting contigs are mapped back to the repeat masked human reference genome, and consensus bases are called from the highest-scoring mapped contigs.











