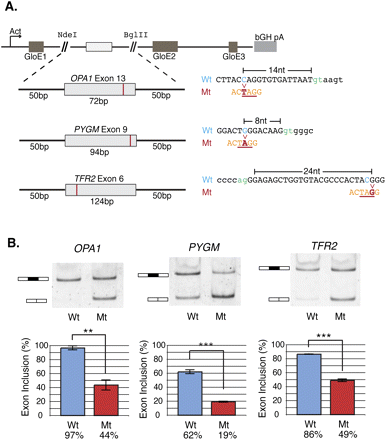
Validation of mutations creating the enriched silencer ACUAGG using the beta-globin splicing reporter. (A) Splicing reporter constructs created from matched pairs of wild-type (Wt) or mutant (Mt) alleles that give rise to a gain of the ACUAGG silencer in constitutive exons in three different disease genes: OPA1, PYGM, and TFR2. GloE1, GloE2, and GloE3 designate exons 1–3 of beta-globin. The polyadenylation signal from the bovine growth hormone 1 gene is indicated by bGH pA. (Blue) Wild-type allele; (red) the mutant; (orange) the silencer sequence created by the mutation. (B) HeLa cells were transiently transfected in triplicate with both wild-type (Wt) and mutant (Mt) alleles. Twenty-four hours after transfection, cells were treated with emetine to inhibit NMD, RNA was harvested, and the splicing efficiency was determined by RT-PCR and visualized using 6% non-denaturing (29:1) polyacrylamide gel electrophoresis (PAGE). The graphs depict mean exon inclusion quantified using an Agilent 2100 Bioanalyzer with standard error bars (see Methods). Statistical hypothesis testing on means was executed using a Welch t-test for normal data with unequal sample size and variance using α-values of (*) 0.05, (**) 0.01, and (***) 0.001.











