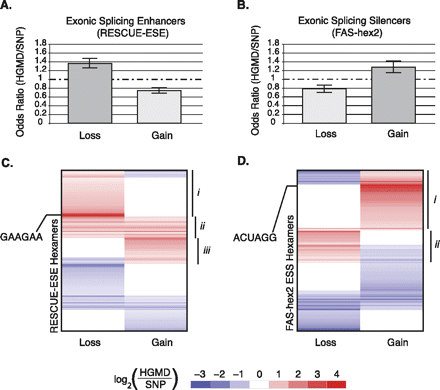
Patterns of exonic splicing regulator loss or gain among pathological mutations (HGMD) as compared to putatively neutral SNPs. (A,B) Bar height corresponds to the odds ratio (OR) of HGMD/SNPs for the loss or gain of enhancers and silencers, respectively. Each error bar represents a two-tailed 95% confidence interval for the bar height (see Methods). Directionality was expressed in the form of the ancestral state > variant for the SNPs and healthy > disease for the HGMD mutations. (A) Hexamers corresponding to exonic splicing enhancers were obtained from the RESCUE-ESE database. Each hexamer was scored for the loss or gain (de novo creation) of an ESE by the inherited disease-causing mutations (relative to the wild-type allele) or putatively neutral SNPs (relative to the ancestral allele). (B) Hexamers corresponding to exonic splicing silencers were obtained from the FAS-hex2 database and scored for loss or gain as described in A. (C,D) Principal component analysis (PCA) of normalized ratios of HGMD versus SNP substitution for loss or gain of ESE and ESS hexamers, respectively. Each row corresponds to a single ESE or ESS hexamer, whereas each column represents loss or gain of the hexamer by a genomic variant. Any hexamers that were not significant at the 5% level were omitted from the heat map. Each box depicts the log ratio for the counts of HGMD/SNP causing loss or gain of a specific hexamer. A positive log ratio in red corresponds to a hexamer in a certain context (column) that is significantly enriched in inherited disease. Alternatively, a blue value represents a hexamer that is polymorphic across human populations. White boxes correspond to non-significant P-values given a false discovery rate (FDR) of 5%. (C) Hexamer clusters corresponding to ESE-loss (region i), ESE-loss and ESE-gain (region ii), and ESE-gain (region iii). Hexamer clusters corresponding to ESS-gain (region i) and ESS-loss (region ii). The loss/gain of SRSF1-like binding sites is indicated by GAAGAA in C, whereas the ACUAGG hexamer is indicated in D.











