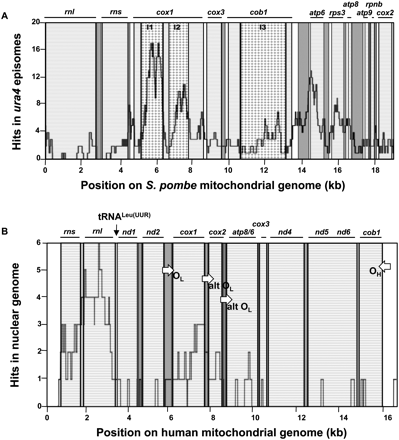
Fragile sites are present in mitochondrial genomes of S. pombe and human. (A) Distribution of S. pombe mtDNA sequences at repair junctions of circularized ura4 fragments. One-hundred-ninety-five mtDNA inserts (83–4004 bp) recovered in ura4 episomes were sequenced. The graph gives the number of times each nucleotide from the S. pombe mitochondrial genome was recovered in episomes. (B) Distribution of mtDNA nucleotides in the 37 human-specific NUMTs described previously (Ricchetti et al. 2004). The graph gives the number of times each nucleotide from the human mitochondrial genome was recovered in human-specific NUMTs. (OH) Heavy-strand synthesis origin; (OL) light-strand synthesis origin; (alt OL) alternative OL. In A and B: (dashed boxes) genes; (gray) tRNAs; (white) intergenic regions. Draughtboards indicate gene introns in S. pombe mitochondrial genome. Position of tRNALeu(UUR) is indicated on human mitochondrial DNA map.











