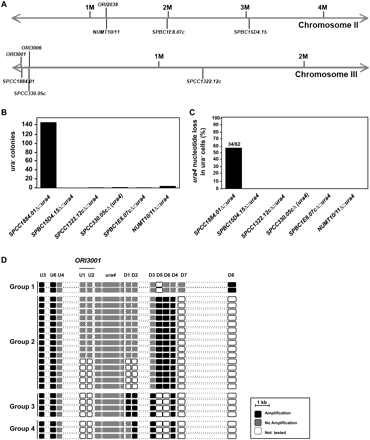
Replication stress induces frequent nucleotide deletion events at subtelomeric ORI3001 locus. (A) An expression cassette comprising the ura4 gene was inserted at five distinct chromosomal loci in S. pombe to give AD549 (SPCC1884.01Δ∷ura4), AD496 (SPBC1E8.07cΔ∷ura4), AD555 (NUMT10/11Δ∷ura4), PN4095 (SPCC1322.12cΔ∷ura4), and AD421 (SPBC15D4.15Δ∷ura4) strains. AD467 strain has an endogenous WT ura4 gene (SPCC330.05c). Positions of ORI3001, ORI3006, and ORI2038 were deduced from the peaks of Mcm6 binding onto chromatin obtained by Hayashi et al. (2007) (Supplemental Fig. 7) and the data from Heichinger et al. (2006). The nomenclature used for ORIs comes from (Heichinger et al. 2006). (B) pREP3x∷cdc18 plasmid was introduced into all six S. pombe strains. Cdc18 overexpression was achieved by incubating cells for 19 h at 32°C in the absence of thiamin. Cells (0.5 × 109) were then plated onto 5-FOA-containing EMM2 agar plates (50 × 106 cells/plate) and the 5-FOAR colonies that were recovered after 4–6 d were tested for uracil auxotrophy. The graph gives the number of ura− colonies that were obtained for each strain. (C) The ura− colonies recovered in B were tested for the presence of the ura4 gene by PCR screen. (D) The extent of genomic DNA deletion was analyzed by PCR in 24 colonies with the ura4 deletion derived from the SPCC1884.01Δ∷ura4 strain. Each box represents a PCR product (see Supplemental Table III for primers). In ura− mutants, PCR products were either recovered (black boxes) or not (gray boxes). White boxes indicate regions that were not tested by PCR. The mutants were classified into four groups according to the extent of genomic DNA deletion. The position of ORI3001 is indicated.











