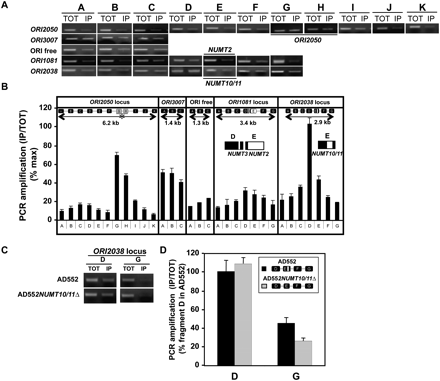
NUMTs are probably not acting as replication origins on their own in S. pombe chromosomes. (A) Lysates from orp1∷orp1-5flag cells (strain AD552) were subjected to ChIP analysis using anti-Flag antibodies. Total chromatin (TOT) or DNA associated with immunoprecipitates (IP) was amplified using primer sets that amplify consecutive ∼300-bp fragments along ORI2050 (ARS2004) (Wuarin et al. 2002) (chromosome II: 1541299–1547496), ORI3007 (chromosome III: 122883–124270), an ORI-free region (chromosome II: 3034667–3035885), ORI1081-NUMT2/3 (chromosome I: 2665851–2669206), and ORI2038-NUMT10/11 (chromosome II: 1242241–1245130). The nomenclature used for ORIs is based on Heichinger et al. (2006). (B) The size of the five regions analyzed and the position of PCR fragments are indicated in each case. PCR reaction products were run on gel and quantified with Image Station 440 CF (Kodak). Band intensity of IP was normalized with TOT for each PCR fragment to give IP/TOT values (100% corresponds to IP/TOT value measured for fragment D of ORI2038 locus). Screening of the ORI2050 genomic locus was used as positive control and revealed Orc1 binding onto fragments G and H (*), as expected (Wuarin et al. 2002). (C,D) The ChIP experiment was repeated in strains AD552 and AD552NUMT10/11Δ (AD566) to test the impact of NUMT10/11 deletion on Orc1 binding onto fragments D and G of ORI2038 locus described in panel A. Data result from two independent experiments and measurements were performed in triplicate. Error bars indicate standard deviations.











