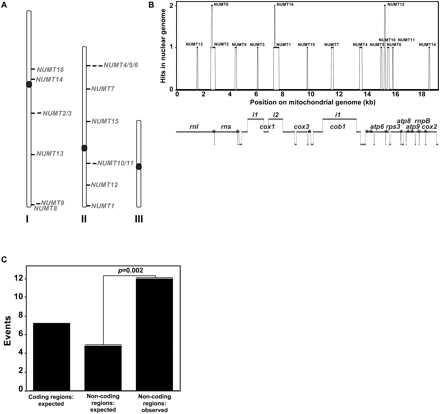
The 16 S. pombe NUMTs are distributed over noncoding regions of chromosomes I and II. (A) Chromosomal mapping of S. pombe single and multiple NUMTs. The total number of independent NUMT insertion sites is estimated at 12. Position of the centromeres is indicated. (B) S. pombe NUMTs are not duplicated. Position of NUMTs is shown on the linear map of S. pombe mitochondrial genome (bottom panel). The graph represents the number of hits in the nuclear genome for the entire mtDNA sequence. (Bottom panel) mtDNA genes are indicated on mtDNA map; i1 and i2 refer to introns; intergenic regions of mtDNA genome are indicated by gray bars downstream from the genes; asterisks indicate the position of tRNAs. (C) The 12 independent NUMT insertion sites from S. pombe were classified according to their genomic localization: coding or noncoding regions. NUMT distribution was compared with the expected frequency based on the contribution of coding and noncoding DNA sequences in S. pombe genome. Fisher's exact probability test was performed on http://faculty.vassar.edu/lowry/odds2x2.html.











