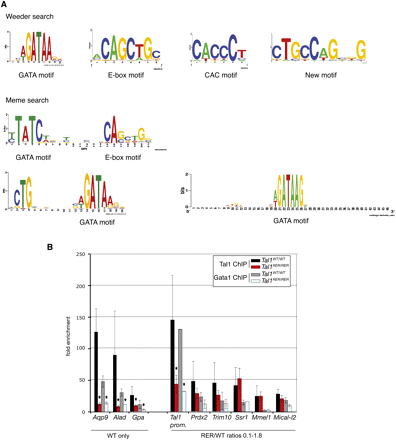
DNA motifs underlying the TAL1 peaks. (A) Logos representing the motifs identified in the sequences underlying the TAL1 peaks using de novo Weeder and Meme searches. (B) Real-time PCR analysis of anti-TAL1 and anti-GATA1 ChIP on selected loci. Chromatin derived from Ter119- populations from Tal1WT/WT and Tal1RER/RER fetal liver culture cells was immunoprecipitated using anti-TAL1 and anti-GATA1 antibodies and the loci indicated on the graph analyzed by real-time PCR. The y-axis represents the enrichment over input DNA, normalized to a control sequence in the Gapdh gene. Error bars, ±1 SD from at least three independent experiments (*P < 0.01). Below the graph are shown the categories the genes belong to, as detected by ChIP-seq.











