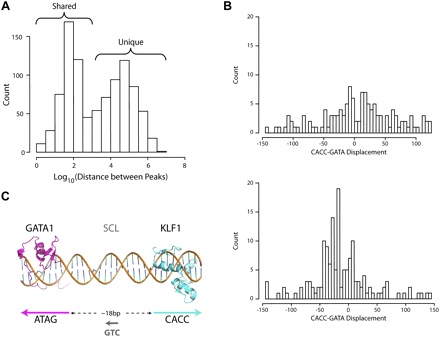
Correlation with GATA1 ChIP-seq reveals extensive co-occupancy by KLF1 and GATA1 throughout the erythroid genome. (A) A histogram of the distance between all KLF1 ChIP-seq peaks and the nearest GATA1 ChIP-seq peak as determined by Cheng et al. (2009) plotted on a Log10 scale as for Figure 1A. The bimodal distribution of distances indicates shared and unique binding sites for KLF1 as shown. (B) Histogram plots of the distance between the KLF1 motif and GATA1 motifs in the same (same strand, top) and opposing (opposite strand, bottom) directions for the KLF1 ChIP-seq peaks. (C) A model for KLF1-GATA1 cooperation based on the data presented in B. Crystal structures of the GATA1 DNA-binding zinc fingers and Zif268 (representative of KLF1) imposed onto a DNA backbone are shown in pink and blue, respectively. The preferred directions and spacing of the motifs for cooperation is shown. The hypothesized existence of SCL between the two factors binding to a “CTG” motif is indicated in gray.











