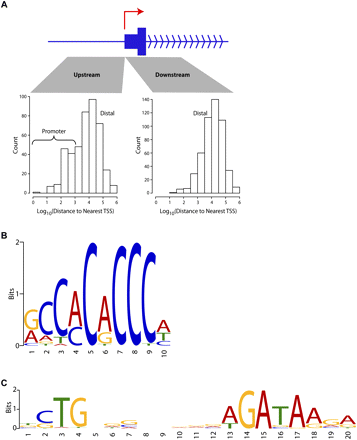
KLF1 ChIP-seq reveals a role in both proximal and distal gene regulation and a refined in vivo binding site motif. (A) Characteristics of KLF1 binding with respect to transcriptional start sites (TSSs). Distances between all KLF1 ChIP-seq peak sites and the nearest mm9 UCSC Genome Browser annotated TSS were determined and sorted into upstream and downstream sets as shown. Distances are plotted as a histogram on a Log10 scale (e.g., 3 represents 1000 bp). (B) KLF1 in vivo consensus binding site predicted by the motif discovery algorithm MEME 4.3 and presented as a sequence LOGO. (C) Analysis of KLF1 ChIP-seq peaks with MEME 4.3 also uncovered the motif shown here which strikingly resembles the motif for murine TAL1(SCL)-GATA1 from the JASPAR CORE database (TOMTOM E-value = 6.6 × 10−11).











