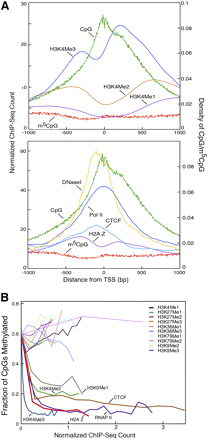
Relationship between DNA methylation, histone modification, chromatin proteins, and nucleosome positioning. (A) m5CpG and CpG densities, ChIP-seq scores, and DNase hypersensitivity scores are plotted relative to promoter TSSs for 16,181 RefSeq genes. (B) CpG methylation plotted as a function of histone modifications, chromatin factors, and RNA polymerase II occupancy. Note the strong negative relationship between DNA methylation and density of H3K4me3 and H2A.Z and the lack of a strong association between DNA methylation status and most histone modifications.











