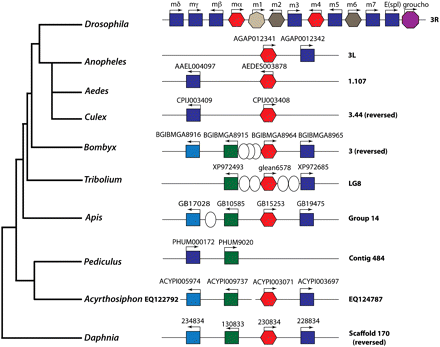
Arthropod E(spl) complexes. Genomic architecture of the arthropod E(spl)-C. E(spl) complexes from Arthropod genome sequences. Names of gene and their respective species are shown in Supplemental Table 1. To the left is a phylogenetic tree of the species represented. (Squares) bHLH genes; (hexagons) bearded class genes; (light gray heptagon) m1; (purple octagon) groucho; (ovals) intervening genes in arthropod E(spl) complexes with no similarity to Drosophila E(spl)-C genes. Genes are color coded with reference to their protein phylogeny: (light blue) E(spl)-C bHLH2-derived sequences; (dark blue) E(spl)-C bHLH1 sequences; (green) Her-derived sequences; (red) Tom/Ocho/bearded-like sequences; (dark gray) m6 sequences. Contigs are shown and labeled to the right. Breaks in the complex are denoted by breaks in the lines representing contigs. For clarity, identifiers for Daphnia genes are missing NCBI_GNO_ prefix.











