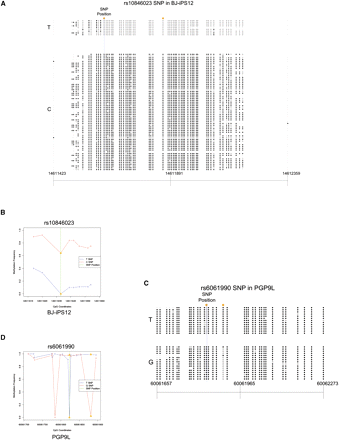
Allele-specific methylation. (A) Sanger reads from BJ-iPS12 show an extended ASM region in dbSNP 129 rs10846023 indexed region (chr12:14611423–14612359) in the intron of the RAB11FIP1 (also known as FLJ22622) gene. This ASM region includes two CpG dinucleotides that overlap with SNPs, but the ASM is not limited to these sites. (Orange circles) SNPs that overlap with CpG dinucleotides. (B) An allele-specific methylation frequency graph based on aligned Illumina data showing ASM at rs10846023 in BJ-iPS12 (chr12:14611616–14611654). (Y-axis) Methylation frequency, where a value of 1 indicates complete methylation at a CpG dinucleotide; (x-axis) chromosomal coordinates. (C) Sanger sequence data around SNP site rs6061990 (TAF4 intronic region, chr20:60061657–60062273) in PGP9L illustrates an example where ASM is solely dependent on SNPs at CpG dinucleotides. (D) An allele-specific methylation frequency graph based on Illumina data showing ASM at rs6061990 in PGP9L (chr20:60061712–60061895). (Green line) SNP position; (orange triangles) SNPs that overlap with CpG dinucleotides. Note that the Illumina data show ASM at a CpG (chr20:60061784) that is not supported by the Sanger data.











