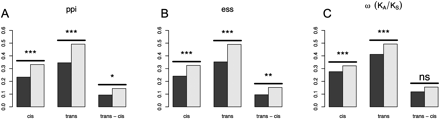
Constraint in expression polymorphism. (A) Genes with presently no detected protein–protein interaction (ppi) partners in current data (light bars) versus genes in the upper 50th percentile of those showing interactions (dark bars). (B) Nonessential genes (light bars) are genes whose homozygous knockouts have a fitness of greater than 0.85. Essential genes (light bars) are those genes where homozygous knockouts are lethal. (C) Genes in the lower 50th percentile of ω (dark bars) versus genes in the upper 50th percentile of ω (light bars). Pairs of bars labeled “cis” and “trans” compare the mean expression divergence |log2(e)| between putatively strongly constrained and weakly constrained categories. The category “trans − cis” compares the quantity |log2(etrans)| − |log2(ecis)| between putatively strongly constrained and weakly constrained categories. Significance is indicated as follows: ns, P > 0.05; *, 0.01 < P < 0.05; **, 0.01 < P < 0.001; ***, P < 0.001. Both cis and trans mutations are subject to purifying selection in all comparisons. trans mutations are more sensitive to changes in constraint than are cis changes when constraint is determined by number of protein interactions or essentiality, but not when measured by protein coding sequence (ω).











