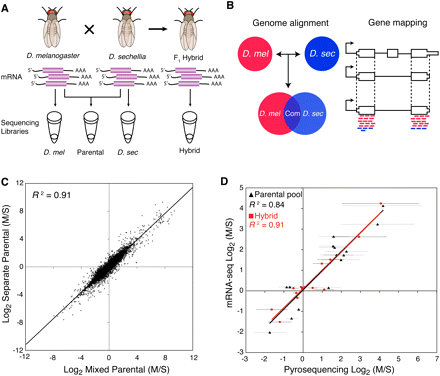
Overview of allele-specific expression profiling by mRNA-seq. (A) mRNA prepared from D. melanogaster, D. sechellia, and their F1 hybrids were used to create four different sequencing libraries: F1 hybrid (Hybrid), mixed parental (Parental), and two species-specific libraries (D. mel and D. sec). (B) Allele-specific sequences were chosen by comparative genomic alignments and mapped to constitutive regions of genes. (C) Comparison of relative gene expression levels (M/S) measured from mixed and separate parental samples with at least 20 mapped reads per gene. (D) Comparison of allele-specific expression ratios determined by mRNA-seq and pyrosequencing. Error bars indicate 95% confidence intervals for pyrosequencing data.











