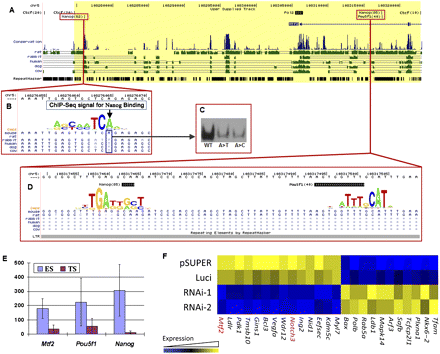
Mouse-specific expression of Mtf2 is due to the creation of transcription factor binding sites produced through the combined effects of a single nucleotide mutation and transposon-introduced sequences. (A) Binding sites for POU5F1, SOX2, NANOG, and RNA Polymerase II lie in close proximity to the mouse Mtf2 gene, between two CTCF binding sites (yellow region). (B) A single nucleotide mutation generates a NANOG binding site in the mouse, but not in human or cow. ChIP-seq data accumulated over 62 sequence reads, overlapping on a single nucleotide in the mouse genome. (C) Electrophoretic mobility shift assay results confirm that mutation of the mouse binding site back to the version in other mammals (A-to-T) eliminates NANOG binding in vitro. (A>T) A-to-T mutation; (A>C) A-to-C mutation. (D) A mouse-specific transposable element inserts a CRM for POU5F1 and NANOG into the region upstream of Mtf2: This CRM is bound by POU5F1 and NANOG in mouse ES cells. (E) Expression levels of Mtf2, Pou5f1, and Nanog in mouse ES and trophoblastic stem (TS) cells. (F) Mtf2 knockdown in mouse ES cells affects the expression of other transcription factors and signaling proteins, including the down-regulation of another ICM-specific gene, Notch3. pSUPER and Luci are control vectors that carry an empty vector (pSUPER) and shRNA against a luciferase gene (Luci). RNAi-1 and RNAi-2 are knockdowns mediated by two different shRNAs.











