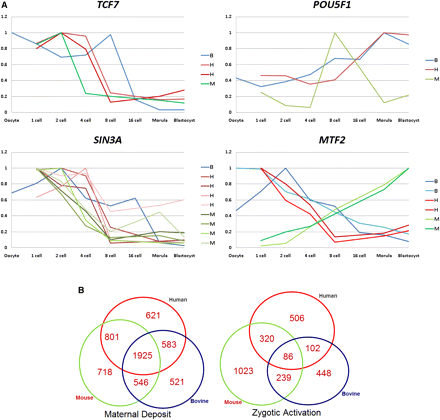
Cross-species comparison of gene expression. (A) Pictorial description of trends in transcription of a subset of genes with distinct roles during PED. Human, mouse, and bovine probe sets are depicted by red, green, and blue curves, respectively. When a gene is targeted by multiple probe sets, all probe sets are shown. The maternally deposited transcript for TCF7 is degraded at two-, four-, and eight-cell stages in mouse, human, and bovine embryos, respectively. SIN3A is highly abundant in zygotes, and exhibits consistent decreasing expression patterns in all three species. POU5F1 exhibits a consistent up-regulation in all three species, however its expression peaked at the eight-cell stage in mouse and at the morula stage in human and bovine. MTF2 transcript exhibits a reversed expression pattern in mouse and in the other two species, as reproduced by two probe sets in each species. (B) Conserved and species-specific MT and ZGAT. A percentage of MT, 33.7%, is shared in all three species, whereas only 3.2% of the ZGAT is shared in all three species. MT has a 10.7-fold larger chance of being shared across species than ZGAT (P-value < 10−30).











