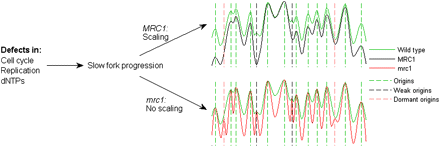
Model of replication timing control. Perturbations of normal replication fork progression (or origin activation), brought about by disruption of replication factors, cause a proportional delay in the activation timing of the different origins, maintaining their relative order of firing. While the structure of the replication timing program is overall maintained, certain origin are rendered inactive because of passive replication. Conversely, in mrc1-deleted cells with a similar disruption of replication, origins are not delayed and the overall structure of the replication program is altered. When combined with the slower progression of the replication fork, this enables the firing of dormant origins, which in wild type are passively replicated. The replication profiles (solid lines) correspond to: green, wild type (unperturbed; normal replication); black, cells subject to a replication perturbation (mutation) with intact MRC1 (scaled replication program); red, cells subject to a replication perturbation with defective MRC1 (no scaling). Broken lines correspond to different classes of replication origins: green, origins active in all situations; black, origins rendered inactive because of scaling when MRC1 is intact and replication is perturbed; red, normally inactive origins that become activated when replication is perturbed and MRC1 is nonfunctional.











