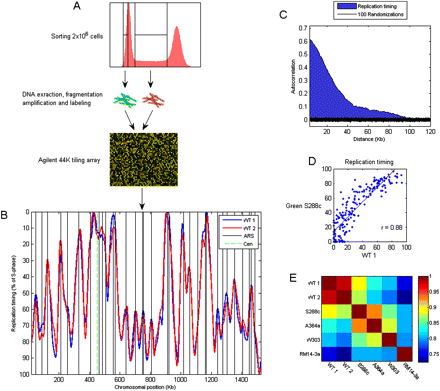
Profiling the genomic DNA replication timing program. (A) Two million SYBR-Green stained cells are sorted separately from G1 and S phases, differentially labeled with fluorescent Cy dyes, and competitively hybridized to an Agilent genomic tiling array of ∼290-bp resolution. The data from four biological replicates are averaged, and the averaged data are smoothed to generate the DNA replication timing profile. (B) The replication profile of chromosome IV generated in two experiments, each consisting of four biological repetitions. The profiles are scaled to 0–100, with 0 being the beginning of S phase and 100 the end. The black vertical lines correspond to positions reported as replication origin (“verified” origins, identified by a combination of experimental and sequence analysis; Nieduszynski et al. 2006). (C) Autocorrelation as a quality control: Shown are the average correlations between the DNA content in chromosomal positions separated by the indicated distances. The correlations are calculated from the raw (unsmoothed) data, are averaged over all chromosomal positions, and are normalized to a maximum of 1. The black region represents 100 separate calculations of autocorrelations on randomized data. (D) Comparison of origin activation times in wild-type cells, as identified in our experiment, with those characterized previously using a similar strain background (S288c [Green et al. 2006]; our strain background is BY4741, a derivative of S288c). See Supplemental Figure S2 for comparison with other data sets. (E) A matrix showing the genome-wide correlations between different data set, including the two repeats from our study. The different experiments used different strain backgrounds, as indicated (S288c and A364a [Green et al. 2006]; W303 [Yabuki et al. 2002]; RM14-3a [Raghuraman et al. 2001]).











