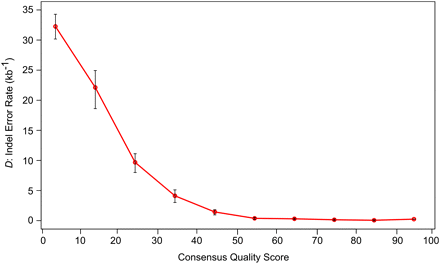
Density of gaps that are errors partitioned by consensus quality score. The Sumatran orangutan genome was divided into 10 bins based upon consensus quality score. For each bin, the indel error rate D was calculated for human-alignable nonrepetitive sequence. Error bars, 95% confidence intervals for the estimation of D. This demonstrates that indel errors occur less commonly in sequence with a high-quality score (Pearson correlation test, P = 0.0038; r = −0.82).











