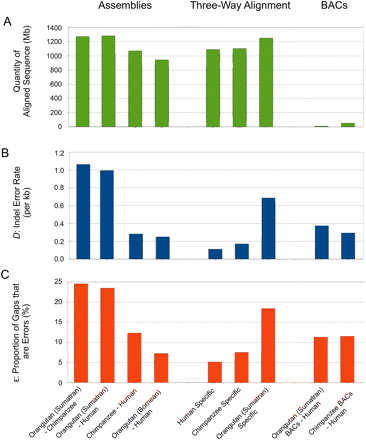
Inferred gap errors are abundant within low coverage regions of the orangutan assembly and are more scarce in both BAC sequence and a hybrid build of capillary sequence (Sumatran) and Illumina (Bornean) orangutan sequence reads. Histograms showing quantities of aligned sequence (A), frequencies of gap errors (B), and proportions of gaps inferred as errors (ɛ) (C), for diverse aligned assemblies. With whole-genome assembly alignments of primates, high error rates are observed for both alignments that contain the Sumatran orangutan assembly. In contrast, the chimpanzee–human alignment contains relatively few errors. When analyzing only the BAC sequences contributed to the Sumatran orangutan assembly, and aligned to human, the indel error rate D is reduced by over twofold. In contrast, alignments between chimpanzee BAC or whole-genome sequence show similar indel error rates. The increased prevalence of gap errors in the Sumatran orangutan assembly is further demonstrated in lineage-specific analysis of a three-way alignment of primate genome assemblies. Analysis of the Bornean build of the orangutan genome using Illumina shotgun reads (fourth column from left) shows a much reduced indel error rate compared with the original Sumatran assembly.











