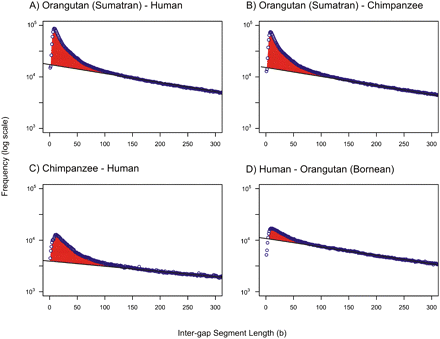
Quantifying gap errors in pairwise alignments of primate genome assemblies. Frequency histograms (natural log scale) of IGS lengths between whole-genome alignments of Sumatran orangutan and human assemblies (A), Sumatran orangutan and chimpanzee assemblies (B), chimpanzee and human assemblies (C), and the human assembly and the Bornean orangutan template assembly (D) created from short reads at 10-fold coverage (see Methods). Repetitive sequence and sequence not placed on chromosomes were excluded (see Methods). Black lines represent the neutral indel model predictions calculated from observed frequencies of IGS lengths (blue circles) between 150 and 300 bases. In all four examples, the expected number of short IGSs is in excess (red) of the number predicted by the neutral indel model. These excesses of short IGSs are due, at least in part, to clusters of gaps representing missing or erroneously inserted sequence, and represent artefacts of the sequencing and assembly process. In alignments of the Sumatran orangutan with human and chimpanzee assemblies, Ng is estimated at 1.3 × 106 and 1.7 × 106, respectively. For alignments of chimpanzee and human, far fewer errors are seen (Ng = 0.3 × 106), suggesting that the anomalies observed in A and B largely reflect inaccuracies in the Sumatran orangutan genome assembly. This is further substantiated by the results for the Bornean orangutan template assembly, which is expected to be more accurate than the Sumatran assembly (Fig. 4).











