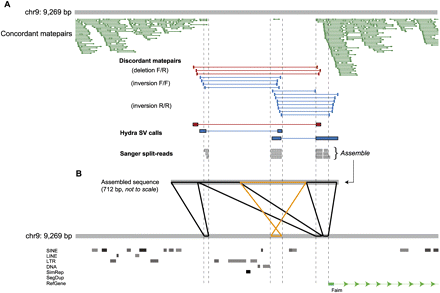
Visualizing a complex SV in a promoter region. (A) A snapshot of aligned sequence data at a validated SV locus from our local mirror of the UCSC Genome Browser (chr9: 98,880,333–98,889,602). At this locus, HYDRA detected one deletion and two inversion breakpoints in the DBA strain from the aligned discordant matepairs (red, those suggesting a deletion; blue, those suggesting an inversion), where F denotes a read mapping to the forward, or plus strand, and R the reverse strand. The dearth of uniquely aligned concordant matepairs (dark green) corroborates the deletion call. Note that a single concordant matepair is aligned within the span of the putative deletion where the two inversion breakpoints overlap, indicating that this segment is not deleted. Three WGS split-reads (gray) from the DBA strain also confirm the HYDRA calls and the observed complexity. (B) The three WGS split-reads were assembled into a 712-bp breakpoint sequence (breaktig) that was then aligned to the reference genome. The image displayed (using PARASIGHT) is representative of the 3316 such images we used to inspect assembled breakpoints. Aligned sections in black are in the same orientation in the breaktig and the reference genome, and the alignments in orange are in the opposite orientation. The complex variant involves two adjacent deletions of 2.5 kb and 0.9 kb, which are separated by an intervening ∼300-bp segment that was not deleted but, rather, inverted. An additional 15-bp deletion is present between the two rightmost alignments to the reference, but is difficult to see at this scale.











