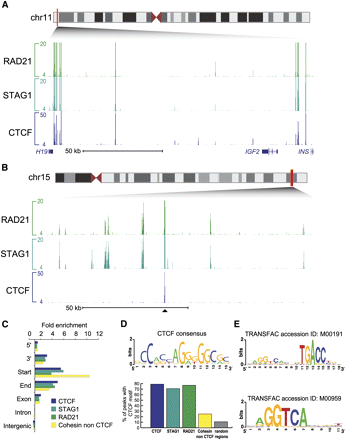
Identification of CTCF-independent cohesin binding events in the human genome. (A) Genomic binding of the cohesin subunits RAD21 and STAG1 as well as CTCF shows colocalization at the H19/IGF2 locus in MCF-7 cells. (B) Illustrative CTCF-independent binding events are shown at a 120-kb region on chromosome 15 in MCF-7 cells; this genomic track is centered around a single shared cohesin-CTCF site. (C) Distribution of cohesin-, CTCF-, and cohesin–non-CTCF-binding (CNC) events in the human genome: 5′, 3′, start, end, exon, intron, and intergenic regions of Ensembl genes are shown. The number of sites was normalized and displayed as fold-enrichment relative to genome background. (D) CTCF consensus DNA motif de novo derived from the MCF-7 CTCF binding events. The occurrence of the CTCF consensus within the CTCF, STAG1, RAD21, CNC events and random genomic regions are presented. (E) Estrogen response elements enriched within the CNC events.











