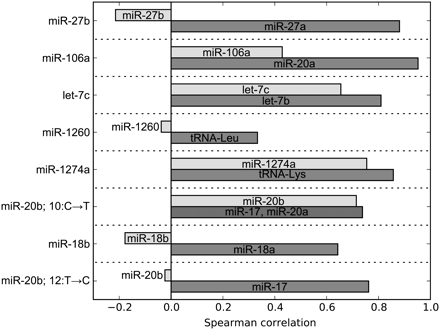
Verification of the cross-mapping correction at spurious editing sites. Our analysis showed that eight out of 10 sites with overrepresented mismatches in mature miRNAs are due to cross-mapping rather than true RNA editing sites (Table 1). For each of these miRNAs, we identified the most abundant RNA that mapped exactly (i.e., without mismatches) to the miRNA locus and the most abundant RNA that mapped to the putative origin of cross-mapping. We also identified the most abundant RNA mapping to the miRNA locus with one mismatch at the site of overrepresented mismatches. For this RNA, we calculated the Spearman correlation along the time course of its counts with the counts of the RNA derived from the miRNA locus (light gray), and the Spearman correlation along the time course of the RNA derived from the putative origin of cross-mapping (dark gray). In all cases, we find that the putative origin of cross-mapping yields a stronger correlation than the miRNA site, supporting our conclusion that the mismatched RNA originates from the cross-mapping locus rather than the miRNA locus.











