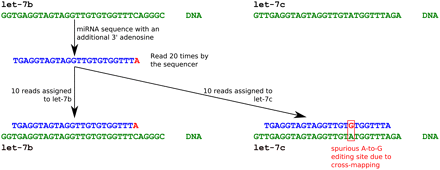
Cross-mapping in short RNA sequencing libraries. Sequences in green and blue represent the genome sequence and the RNA sequence, respectively. Mismatches between the genome sequence and the RNA sequence are shown in red. A miRNA sequence with an additional 3′ adenosine, either due to post-transcriptional addition (as discussed in the text) or a sequencing error, maps equally well to the genome loci encoding the human miRNAs let-7b and let-7c. This miRNA sequence was read 20 times in the FANTOM4 time course short RNA libraries. Dividing the sequence counts equally between these two genome loci leads to a spurious RNA editing site in let-7c. Alternatively, cross-mapping to unannotated genome regions may give rise to spurious novel noncoding RNA loci.











