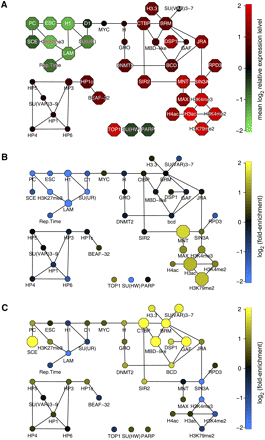
Examples of compartmentalization of regulatory function in the chromatin network. (A) BN80 with the same layout as in Figure 2, with nodes colored according to the mean expression level (in Kc cells) of the target genes of each chromatin component. (B,C) Same as in A, but node colors depicting enrichment (yellow) or depletion (blue) of genes that are expressed in embryonic trunk mesoderm (B) or yolk cells (C). Node sizes in A–C depict the statistical significance of the observed expression level (A, two-sided Wilcoxon test) or the observed enrichment or depletion (B,C, two-sided binomial test), ranging from P > 10−3 (smallest nodes) to P ≤ 10−8 (largest nodes). Tissue expression data in B and C are from (Tomancak et al. 2007).











