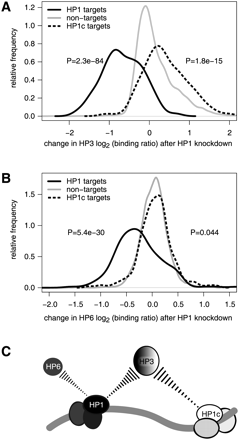
Experimental validation of predicted HP3 and HP6 targeting interactions. Density plots (“smoothed histograms”) showing the changes in binding levels of HP3 (A) and HP6 (B) after RNAi knockdown of HP1, compared to a control knockdown. Changes in binding are shown for subsets of genes as indicated. The y-axes show relative frequency (plots in each panel are normalized to have the same surface area). Binding data after knockdown are from (Greil et al. 2007). Nontargets are bound by neither HP1 nor HP1C. P-values are according to two-sided Wilcoxon tests. (C) Cartoon depicting the competitive targeting of HP3 by HP1 and HP1C, as well as the targeting of HP6 by HP1.











